phylogenetic tree maker from table
In the Run Tool Phylogenetic Tree Builder Tool dialog select alignment if not selected by default and select Distance Method Poisson protein Kimura protein or Jukes Cantor DNA Tree Construct Method Neighbor Joining or Fast Minimum Evolution and Labels for Leaf Nodes representation and click Finish. Or it can be built from molecular information like genetic sequences.

The Mitochondrial Dna Mtdna Haplogroup H Hv Dna Genetics Dna Dna History
Choose Taxonomy Links from the Display pull down list at the top of the results.

. Multiple cladogram templates to quickly start analyzing how groups of organisms are related to each other. Once you have linked together all the species your phylogenetic tree is now ready. Basic phylogenetic functions DrawNewickTreetree Parses a Newick tree string and renders it graphically as a tree with the tips and nodes labeled and the branches drawn proportional to the branch lengths in the file.
Extensive shape libraries for over 50 types of diagrams including food webs dichotomous keys phylogenetic trees and more. STEP 1 - Enter your multiple sequence alignment. Tree annotation made easy.
Parentheses create a parent. Compose speech audio from IPA phonetic transcriptions using Eckher IPA to Speech. Default Clustal Distance Matrix NEXUS.
Simply select any alignment in Geneious Prime and your choice of algorithm to generate your phylogenetic tree with simple one click methods. Or upload a file. Complete clades can be simply included with interruption at desired taxonomic levels and with optional filtering of unwanted nodes.
Data can be imported from many tree formats tables and BayesTraits output. Simple to use drag and drop tools to easily visualize phylogenetic relationships and create cladograms effortlessly. Enter or paste a multiple sequence alignment in any supported format.
To build phylogenetic trees statistical methods are applied to determine the tree topology and calculate the branch lengths that best describe the phylogenetic relationships of the aligned sequences in a dataset. The page will re-load with a Common Tree display. I made this video below the fold to illustrate the steps involved in making a phylogenetic tree.
Manage and visualize your trees directly in the browser and annotate them with various datasets. Individual nodes in the tree link to the Taxonomy. To build phylogenetic trees statistical methods are applied to determine the tree topology and calculate the branch lengths that best describe the phylogenetic relationships of the aligned.
In general the more information youre. The Common Tree display shows a hierarchical view of the relationships among the taxa and their lineages. From a list of taxonomic names identifiers or protein accessions phyloT will generate a pruned tree in the selected output format.
The tree is in the phastCons or nh format namelength. The problem is that I dont have sequences to align yet this is gonna be a part of my project. A phylogenetic tree can be built using physical information like body shape bone structure or behavior.
Root is the common ancestor of the species under study. Annotate your trees directly from Microsoft Excel LibreOffice or Google Sheets or use the integrated web. About Press Copyright Contact us Creators Advertise Developers Terms Privacy Policy Safety How YouTube works Test new features Press Copyright Contact us Creators.
Give the best result and the most informative tree. Length is not required if use-branch-lengths is not checked. In building a tree we organize species into nested groups based on shared derived traits traits different from those of the groups ancestor.
Now the tree is ready and can be saved or shared with everyone. T-REX includes several popular bioinformatics applications such as MUSCLE MAFFT Neighbor Joining NINJA BioNJ PhyML RAxML random phylogenetic tree generator and. As long as you have something you can compare across different species you can make a phylogenetic tree.
Choose Common Tree from the Display pull down menu. T-Rex Tree and reticulogram REConstruction - is dedicated to the reconstruction of phylogenetic trees reticulation networks and to the inference of horizontal gene transfer HGT events. Fig5 a shows an unrooted tree of species A B C and D.
TreeGraph 2 is a is graphical editor for phylogenetic trees which allows to apply various of graphical formats and edit operations and supports several visible or invisible annotations attached to nodes or branches. The basic steps are to. There are actually a lot of different ways to make these trees.
It supports both the 18 column IUPAC and 32 column long form versions of the periodic table and provides the mobile- and touch-friendly interface for viewing the table. An evolutionary tree is a visual demonstration of the evolution of species from its point of origin. The interactive distance matrix viewer allows you to rapidly calculate meaningful statistics for phylogenetics analysis.
A key feature is the interactive comparison and combination of alternative. Create sequence logos for protein and DNARNA alignments using Eckher Sequence Logo Maker. This function requires branchlengths.
Interactive Tree Of Life is an online tool for the display annotation and management of phylogenetic and other trees. Im using about 10 to 15 species across most kingdoms and I was wondering if there is a database. Add colors symbols or fonts from EdrawMaxs library to help differentiate between the linked organisms.
STEP 2 - Set your Phylogeny options. By sporte on March 27 2008. The length of the root branch is usually not specified.
Build a data set. It is a branching representation that portrays a cladistic. Most phylogenetic methods do not locate the root of a tree and the unrooted trees only reflect the relationship among species but not the evolutionary path.
PhyloT generates phylogenetic trees based on the NCBI taxonomy or Genome Taxonomy Database. The sequences of genes or proteins can be compared among. Align sequences build and analyze phylogenetic trees using your choice of algorithm.
A phylogenetic tree is a diagrammatic representation of the development of biological species. Parents must have two children. The most common computational methods applied include distance-matrix methods and discrete data methods such as maximum parsimony.
A phylogeny is a branch of Biology that specially deals with Phylogenesis. A phylogenetic tree may be built using morphological body shape biochemical behavioral or molecular features of species or other groups. Tree is a string containing a tree with branch lengths in Newick format or a.
Use a example sequence Clear sequence See more example inputs.

Infographic The History Of Audio Equipment Audio Pop Chart Infographic

Family Tree Art Family Tree Genealogy Art
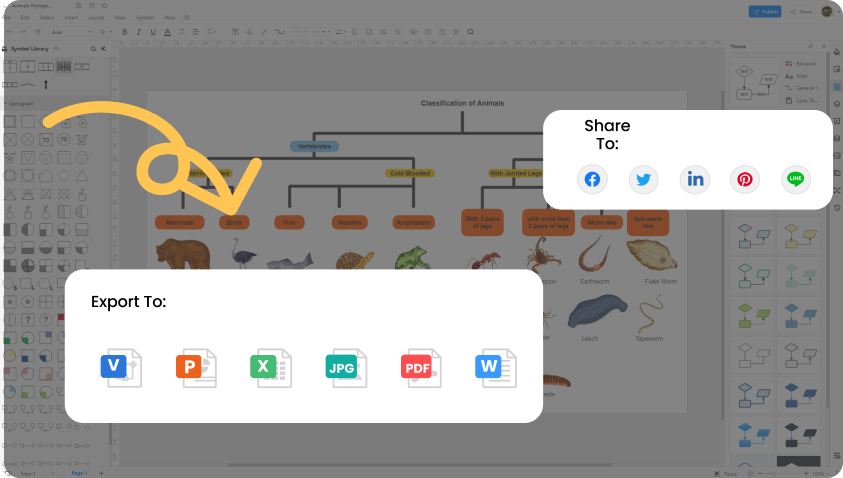
Free Online Phylogenetic Tree Maker Edrawmax Online

Visualizing Family Trees Family Tree Genealogy Tree Tree

Haplogroup H Subclades Phylogenetic Tree Geneaology Genealogy
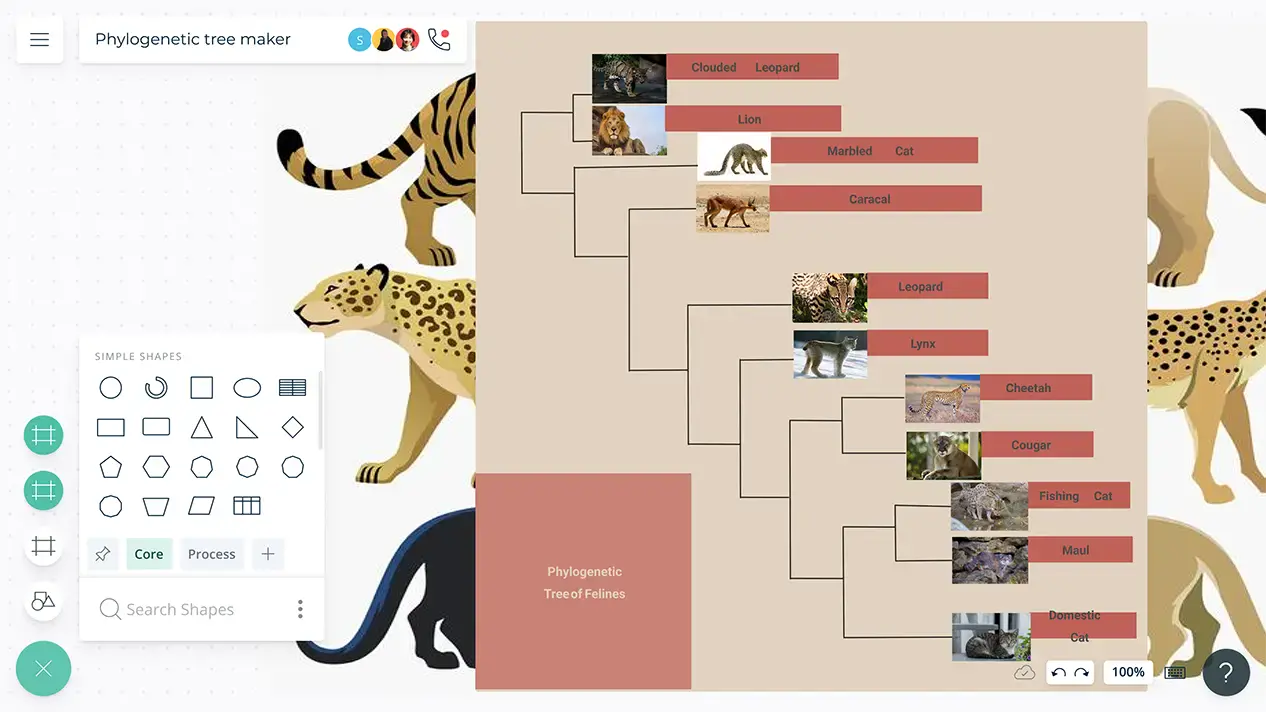
Phylogenetic Tree Maker Phylogenetic Tree Template Creately

Phylogenetic Tree Using Upgma Method Download Scientific Diagram

I Have My Family Tree Back To Adam And Eve Familysearch Blog Family Tree Art Family Tree Genealogy Art

4 Best Free Online Phylogenetic Tree Maker Services

What Is Spirogyra Marine Biology Biology Botany
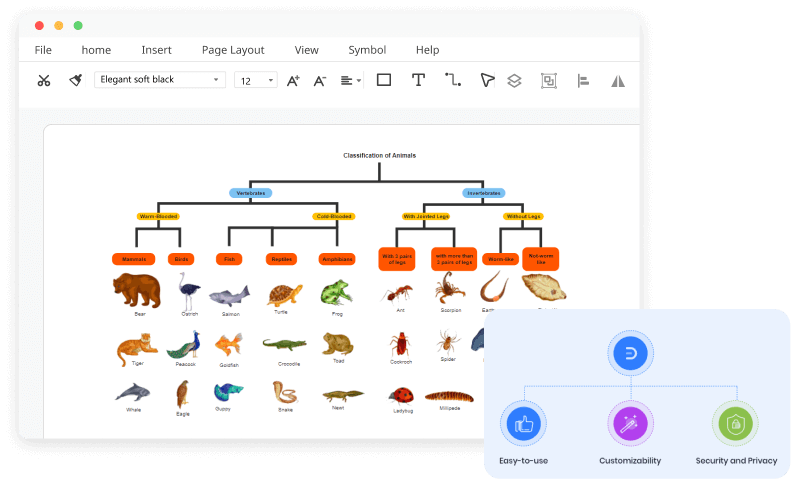
Free Online Phylogenetic Tree Maker Edrawmax Online

Pin De Ricardo Bandeira Em Biomol Imunologia Genetica
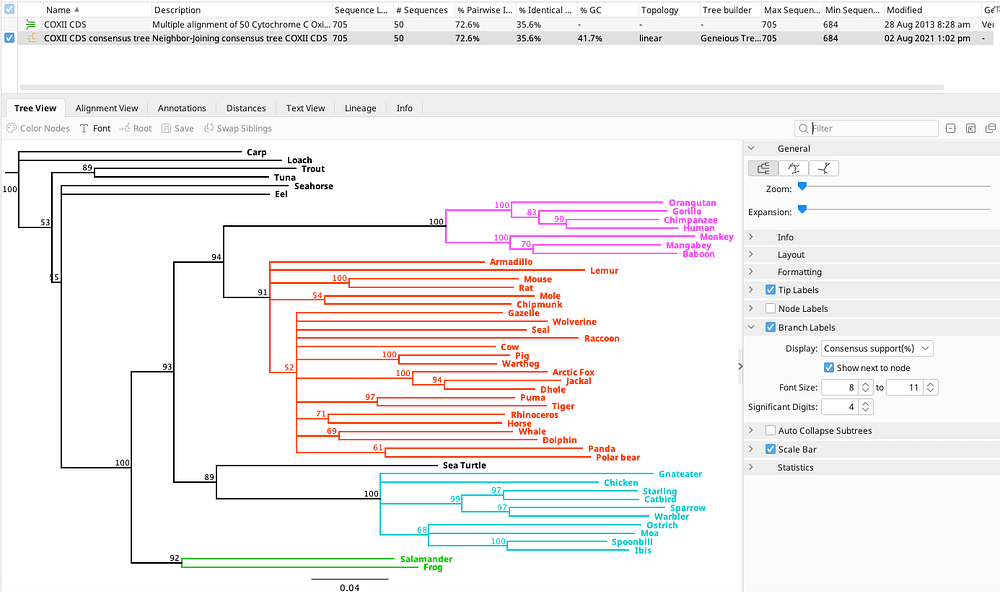
Phylogenetic Tree Building Geneious Prime
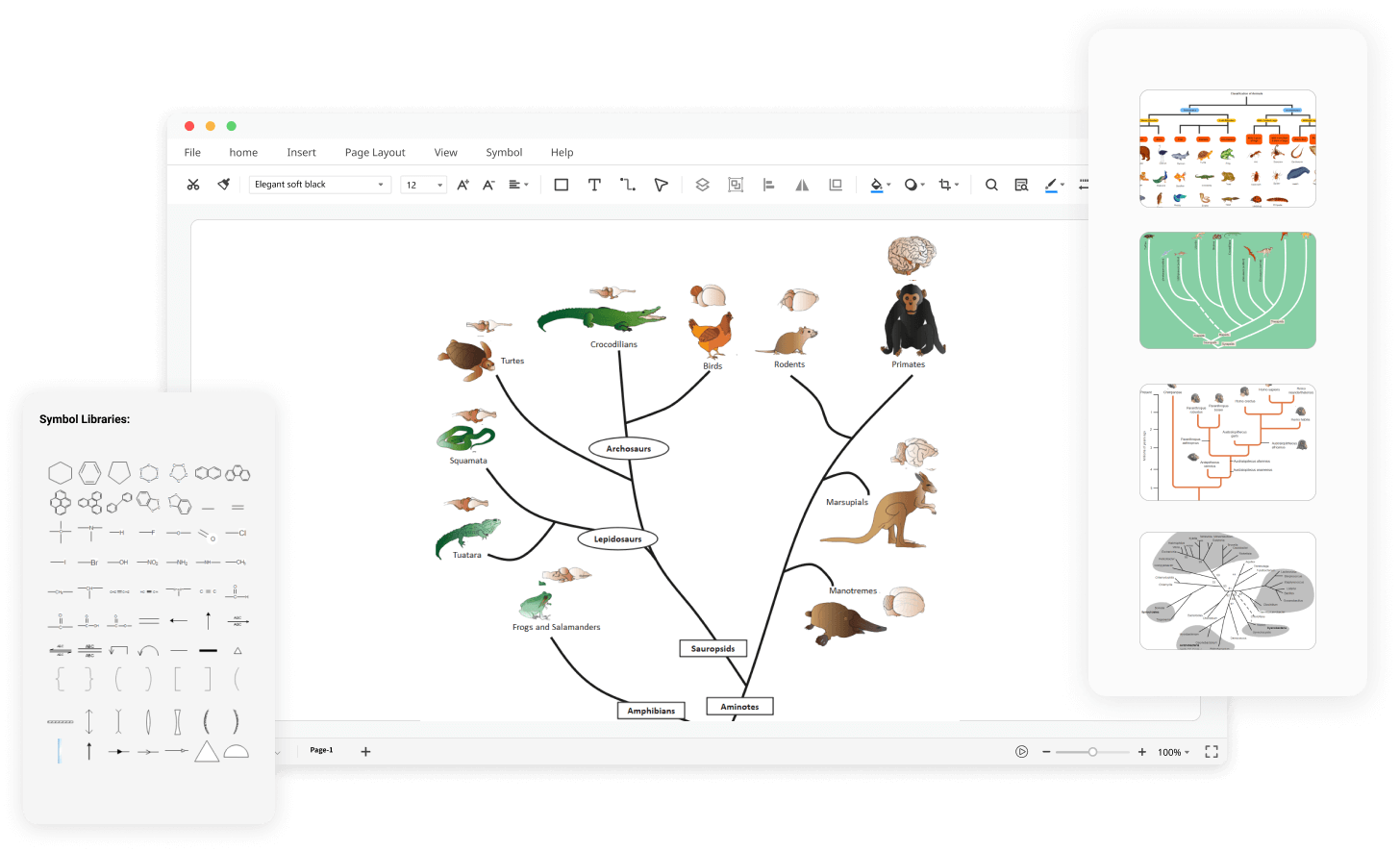
Free Online Phylogenetic Tree Maker Edrawmax Online
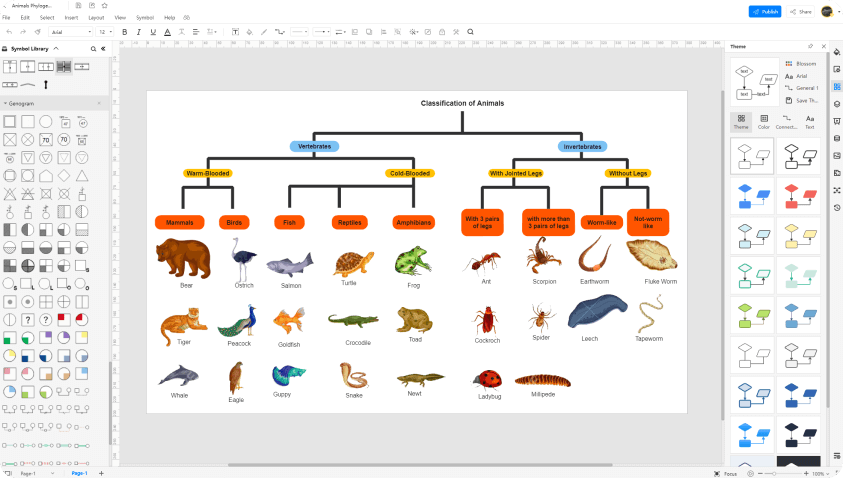
Free Online Phylogenetic Tree Maker Edrawmax Online

Making Phylogenetic Trees From Simple Data Tables Youtube

170 Anos De Reproductores Musicales En Una Sola Imagen Audio Pop Chart Infographic